processing-nirs-with-mne.md 6.5 KB
Reading and processing NIRS data with MNE-Python
This tutorial demonstrates how to read and process NIRS data using
MNE-Python and almirah.
Setup
First, we'll import the necessary libraries and set up the logging and warning configurations.
import mne
import warnings
import numpy as np
import matplotlib.pyplot as plt
from itertools import compress
from almirah import Layout
mne.set_log_level(False)
warnings.filterwarnings('ignore')
Loading the Data
Next, we'll set up the layout to access the NIRS data.
lay = Layout(root="/path/to/data", specification_name="bids")
print(lay)
This should output:
<Layout root: '/path/to/data'>
We can query the layout to find all NIRS files with the .snirf extension:
files = lay.query(datatype="nirs", extension=".snirf")
len(files)
This gives the total number of NIRS files:
1315
Querying a Specific File
To query a specific file, we can filter by subject, task, datatype, and extension:
file = lay.query(subject="D0019", task="rest", datatype="nirs", extension=".snirf")[0]
print(file.rel_path)
This should output the relative path of the file:
sub-D0019/ses-111/nirs/sub-D0019_ses-111_task-rest_run-01_nirs.snirf
We can then download the NIRS file:
file.download()
Reading Raw Data
Next, we read the raw NIRS data:
raw = mne.io.read_raw_snirf(file.path)
raw.load_data()
<summary><strong>General</strong></summary>
<table class="table table-hover table-striped table-sm table-responsive small">
<tr>
<th>Measurement date</th>
<td>November 12, 1917 00:00:00 GMT</td>
</tr>
<tr>
<th>Experimenter</th>
<td>Unknown</td>
</tr>
<tr>
<th>Participant</th>
<td>mne_anonymize</td>
</tr>
</table>
</details>
<details open>
<summary><strong>Channels</strong></summary>
<table class="table table-hover table-striped table-sm table-responsive small">
<tr>
<th>Digitized points</th>
<td>21 points</td>
</tr>
<tr>
<th>Good channels</th>
<td>36 fNIRS (CW amplitude)</td>
</tr>
<tr>
<th>Bad channels</th>
<td>None</td>
</tr>
<tr>
<th>EOG channels</th>
<td>Not available</td>
</tr>
<tr>
<th>ECG channels</th>
<td>Not available</td>
</tr>
</table>
</details>
<details open>
<summary><strong>Data</strong></summary>
<table class="table table-hover table-striped table-sm table-responsive small">
<tr>
<th>Sampling frequency</th>
<td>15.62 Hz</td>
</tr>
<tr>
<th>Highpass</th>
<td>0.00 Hz</td>
</tr>
<tr>
<th>Lowpass</th>
<td>7.81 Hz</td>
</tr>
<tr>
<th>Filenames</th>
<td>sub-D0019_ses-111_task-rest_run-01_nirs.snirf</td>
</tr>
<tr>
<th>Duration</th>
<td>00:10:03 (HH:MM:SS)</td>
</tr>
</table>
</details>
print(raw.info)
<Info | 9 non-empty values
bads: []
ch_names: S1_D1 760, S1_D1 850, S1_D2 760, S1_D2 850, S2_D2 760, S2_D2 ...
chs: 36 fNIRS (CW amplitude)
custom_ref_applied: False
dig: 21 items (3 Cardinal, 18 EEG)
highpass: 0.0 Hz
lowpass: 7.8 Hz
meas_date: 1917-11-12 00:00:00 UTC
nchan: 36
projs: []
sfreq: 15.6 Hz
subject_info: 4 items (dict)
>
Preprocessing
We pick the NIRS channels, compute the source-detector distances, and filter out channels with a distance greater than 0.01:
picks = mne.pick_types(raw.info, meg=False, fnirs=True)
dists = mne.preprocessing.nirs.source_detector_distances(raw.info, picks=picks)
raw.pick(picks[dists > 0.01])
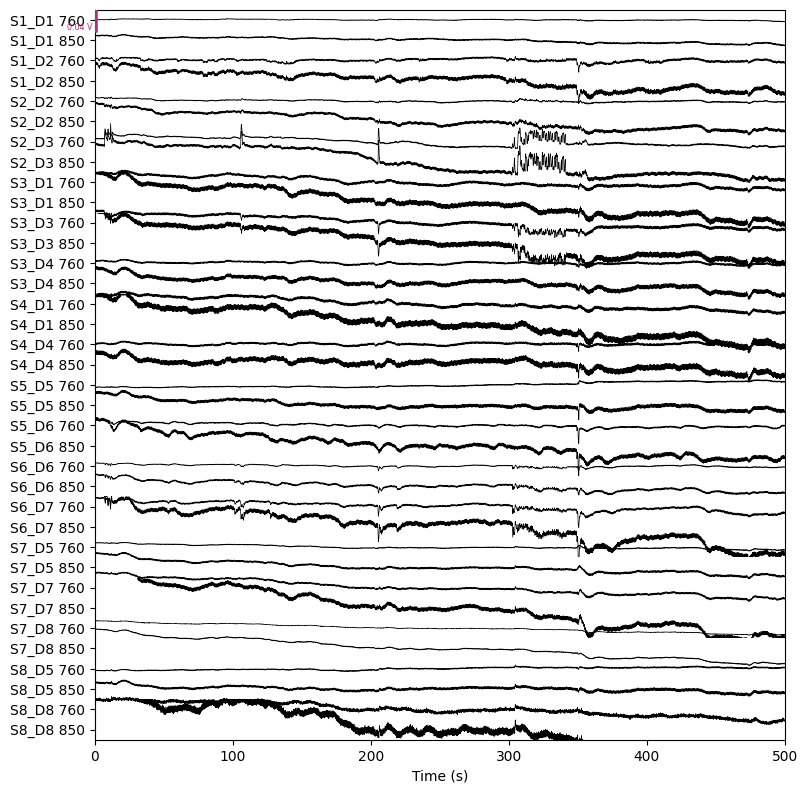
raw.plot(n_channels=len(raw.ch_names), duration=500, show_scrollbars=False)
plt.show()
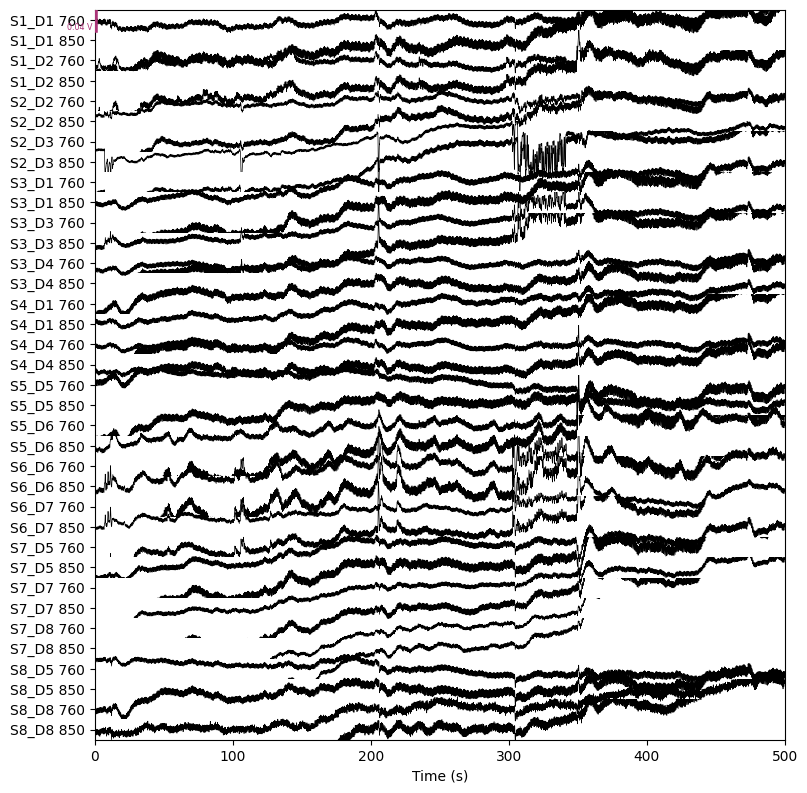
We convert the raw data to optical density:
raw_od = mne.preprocessing.nirs.optical_density(raw)
raw_od.plot(n_channels=len(raw_od.ch_names), duration=500, show_scrollbars=False)
plt.show()
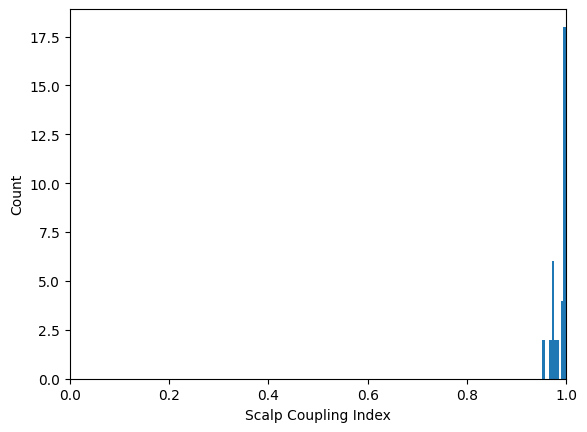
Next, we compute the Scalp Coupling Index (SCI) and plot the distribution:
sci = mne.preprocessing.nirs.scalp_coupling_index(raw_od)
fig, ax = plt.subplots()
ax.hist(sci)
ax.set(xlabel="Scalp Coupling Index", ylabel="Count", xlim=[0, 1])
plt.show()
We mark channels with a low SCI as bad:
raw_od.info["bads"] = list(compress(raw_od.ch_names, sci < 0.5))
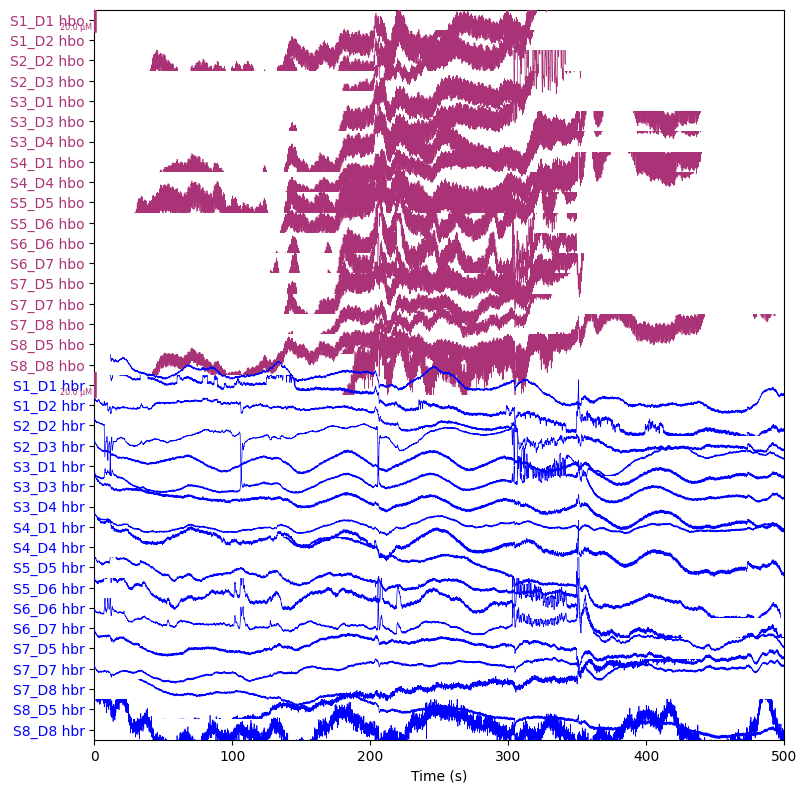
We convert the optical density data to hemoglobin concentration:
raw_haemo = mne.preprocessing.nirs.beer_lambert_law(raw_od, ppf=0.1)
raw_haemo.plot(n_channels=len(raw_haemo.ch_names), duration=500, show_scrollbars=False)
plt.show()
Power Spectral Analysis
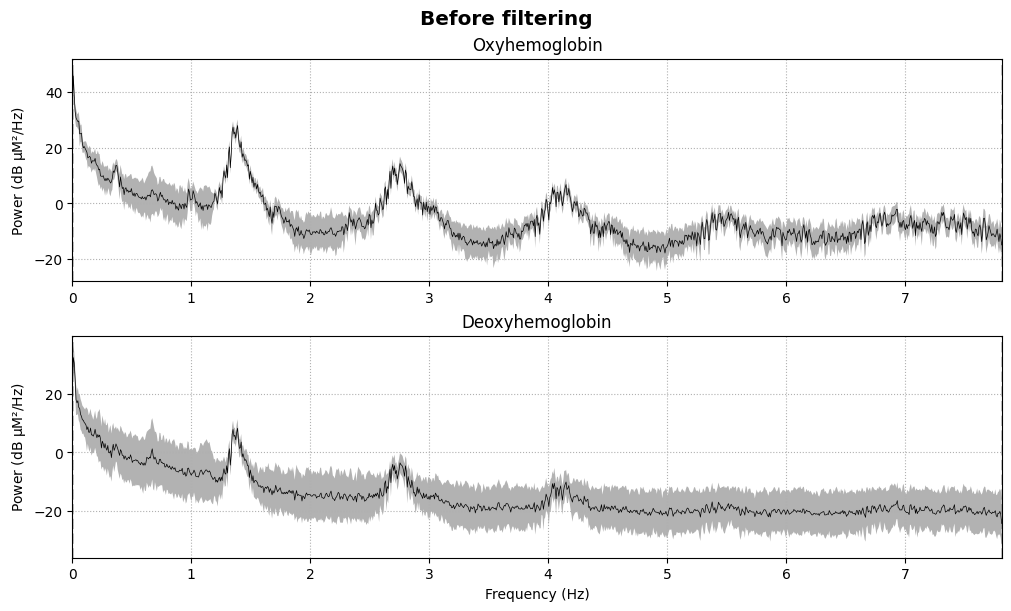
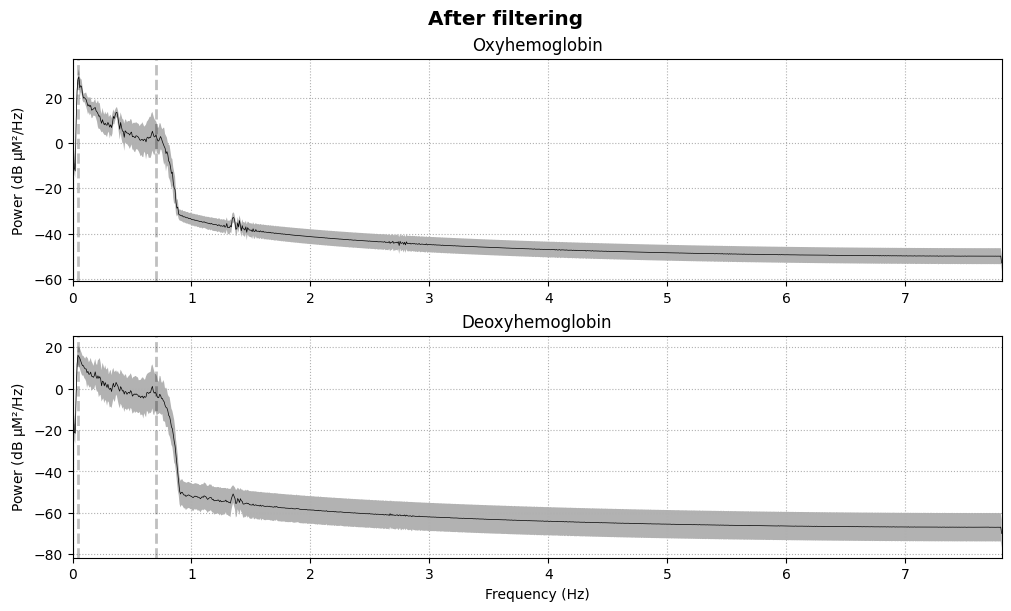
We compute and plot the Power Spectral Density (PSD) before and after filtering:
fig = raw_haemo.compute_psd().plot(average=True, amplitude=False)
fig.suptitle("Before filtering", weight="bold", size="x-large")
raw_haemo = raw_haemo.filter(0.05, 0.7, h_trans_bandwidth=0.2, l_trans_bandwidth=0.02)
fig = raw_haemo.compute_psd().plot(average=True)
fig.suptitle("After filtering", weight="bold", size="x-large")
plt.show()
Epoching
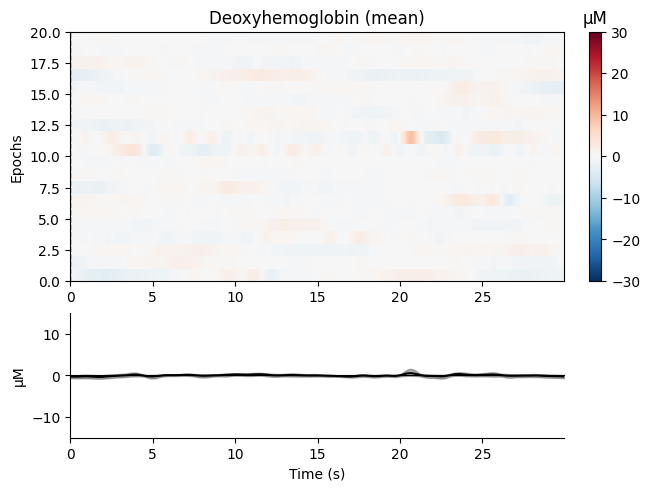
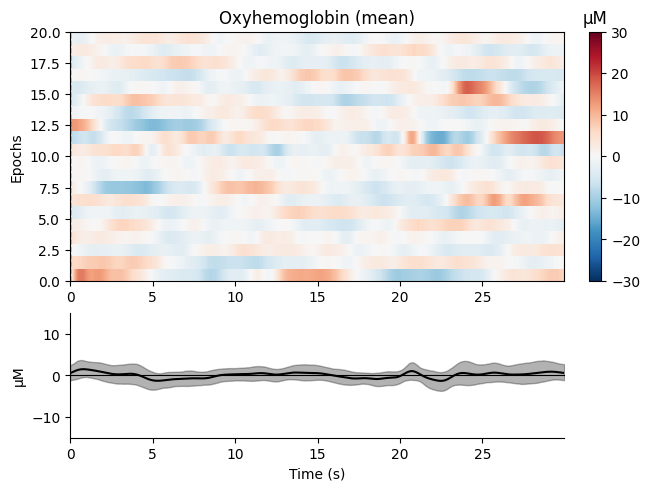
We create epochs from the continuous data:
epochs = mne.make_fixed_length_epochs(raw_haemo, duration=30, preload=False)
Finally, we plot the epochs:
epochs.plot_image(combine="mean", vmin=-30, vmax=30,
ts_args=dict(ylim=dict(hbo=[-15, 15],
hbr=[-15, 15])))
plt.show()
This concludes the tutorial. You've learned how to read, and process
near-infrared spectroscopy data using MNE-Python and almirah.