processing-eeg-with-mne.md 3.8 KB
Processing EEG with MNE-Python
import mne
import warnings
import numpy as np
import matplotlib.pyplot as plt
from almirah import Layout
mne.set_log_level(False)
warnings.filterwarnings('ignore')
lay = Layout(root="/path/to/data", specification_name="bids")
lay
<Layout root: '/path/to/data'>
lay = Layout.get(specification_name='bids')
files = lay.query(datatype="eeg", extension=".vhdr")
len(files)
2223
vhdr_file = lay.query(subject="D0019", session="101", datatype="eeg", task="rest", extension =".vhdr")[0]
eeg_file = lay.query(subject="D0019", session="101", datatype="eeg", task="rest", extension=".eeg")[0]
montage_file = lay.query(subject="D0019", session="101", space="CapTrak", suffix="electrodes")[0]
print(vhdr_file.rel_path)
sub-D0019/ses-101/eeg/sub-D0019_ses-101_task-rest_run-01_eeg.vhdr
eeg_file.download()
montage = mne.channels.read_custom_montage(montage_file.path)
raw = mne.io.read_raw_brainvision(vhdr_file.path, preload=True)
raw.set_montage(montage)
raw.info["bads"] = ["VREF"]
raw.info
<summary><strong>General</strong></summary>
<table class="table table-hover table-striped table-sm table-responsive small">
<tr>
<th>Measurement date</th>
<td>Unknown</td>
</tr>
<tr>
<th>Experimenter</th>
<td>Unknown</td>
</tr>
<tr>
<th>Participant</th>
<td>Unknown</td>
</tr>
</table>
</details>
<details open>
<summary><strong>Channels</strong></summary>
<table class="table table-hover table-striped table-sm table-responsive small">
<tr>
<th>Digitized points</th>
<td>132 points</td>
</tr>
<tr>
<th>Good channels</th>
<td>128 EEG, 5 misc</td>
</tr>
<tr>
<th>Bad channels</th>
<td>VREF</td>
</tr>
<tr>
<th>EOG channels</th>
<td>Not available</td>
</tr>
<tr>
<th>ECG channels</th>
<td>Not available</td>
</tr>
</table>
</details>
<details open>
<summary><strong>Data</strong></summary>
<table class="table table-hover table-striped table-sm table-responsive small">
<tr>
<th>Sampling frequency</th>
<td>1000.00 Hz</td>
</tr>
<tr>
<th>Highpass</th>
<td>0.00 Hz</td>
</tr>
<tr>
<th>Lowpass</th>
<td>500.00 Hz</td>
</tr>
</table>
</details>
Preprocessing
# Apply a band-pass filter
raw.filter(1, 40, fir_design="firwin")
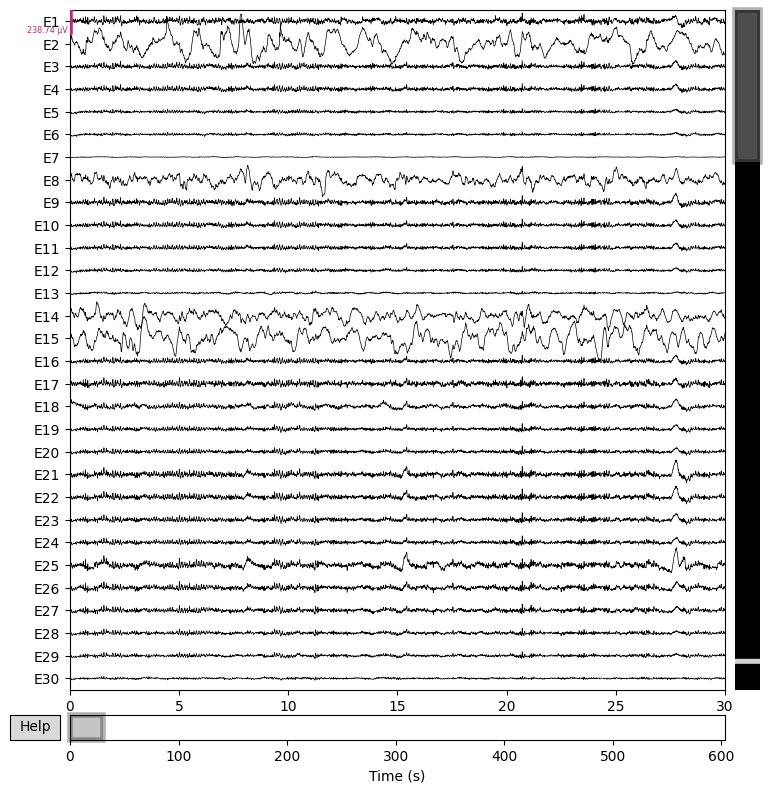
# Plot the raw data
raw.plot(n_channels=30, duration=30, scalings="auto")
plt.close()
Artifact removal using ICA
# Setup ICA
ica = mne.preprocessing.ICA(n_components=15, random_state=97)
ica.fit(raw)
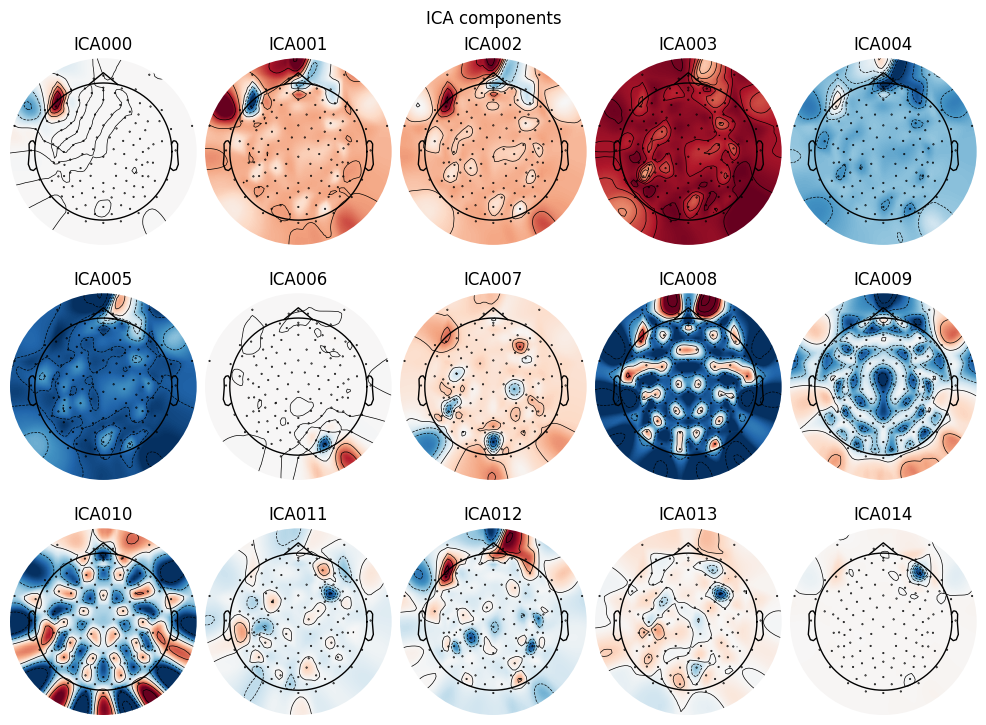
# Plot ICA components
ica.plot_components()
# Select bad components manually (usually by visual inspection)
ica.exclude = [0, 6, 14] # Example: components 0 and 1 are eye blinks and heartbeats
# Apply ICA removal
raw_clean = ica.apply(raw.copy())
Power Spectral Analysis
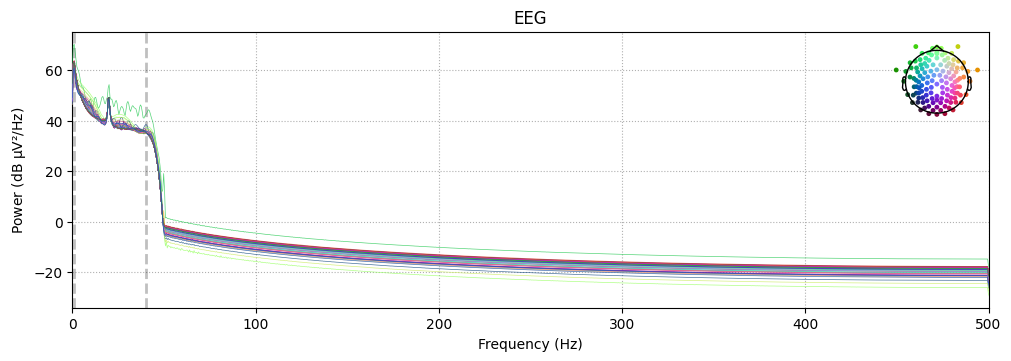
# Compute and plot Power Spectral Density (PSD)
raw_clean.compute_psd().plot(exclude="bads", amplitude=False)
plt.show()
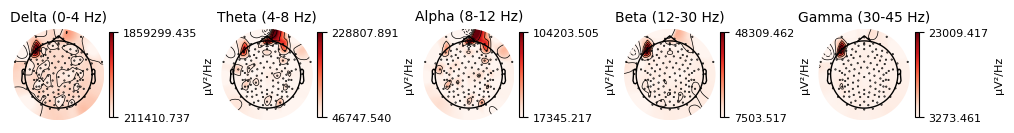
raw_clean.compute_psd().plot_topomap(ch_type="eeg", agg_fun=np.median)
plt.close()